Query submission
Query tab and input data
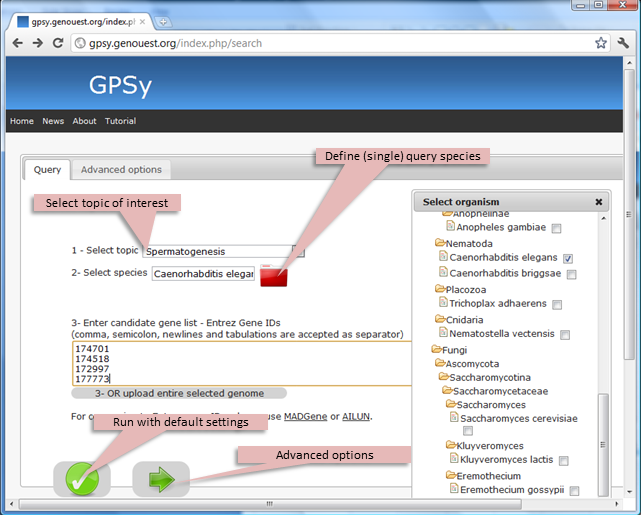
The interface includes a Query tab which enables users to:
- 1 - select one biological Topic from a dropdown-menu
- 2 - define the query species by clicking on the red folder (H. sapiens is the default setting; you can choose only a single species for your query)
- 3 - Enter the list of candidates into the text field
Alternatively, the user can request prioritization of the entire genome for the chosen species using the Upload entire selected genome option. Currently, GPSy only accepts Entrez Gene identifiers (IDs) because reliable and consistent gene ID conversion is a complex problem; users are referred to two up-to-date resources for gene ID unification over a wide range or organisms, MADGene and AILUN.
Advanced options tab
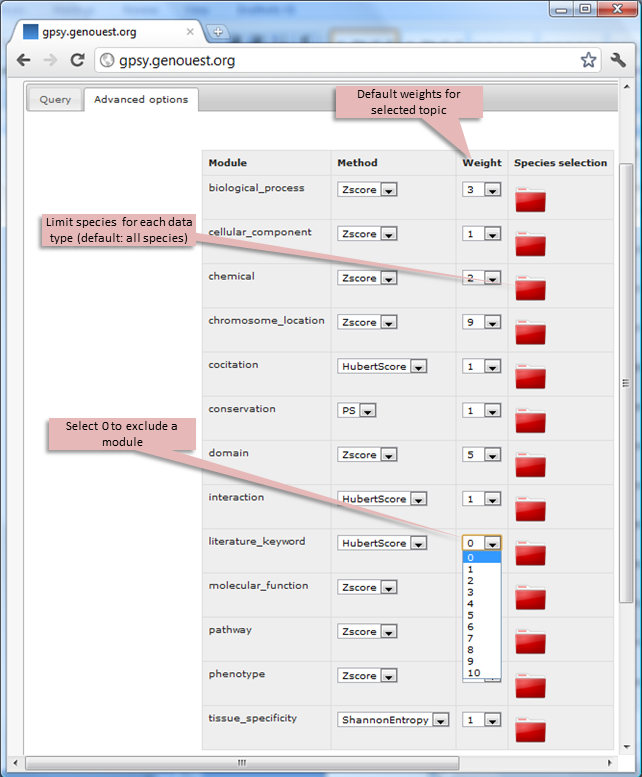
It is possible to select individual species and data modules and to modify their weights (from 0 to 10) using the Advanced options tab. By default, all datasets are selected for all species and the preset parameters from the optimal weight scheme are applied.
Results and output data
The output page displays the top 50 genes by default but users can change this setting as they deem appropriate.
The result is displayed in the form of a table containing one gene per line with columns containing the Entrez Gene ID (hyperlinked to the NCBI), the gene symbol (Description), the rank (Priority list), the number of modules used to compute the rank (NB_values), the overall score (Score), the initial position of the genes in the uploaded list (Initial order), followed by individual module ranks in alphabetical order (Biological process, Cellular component, Cocitation, Conservation, Domain, Interaction, Molecular function, Pathway, Phenotype, Tissue specificity).
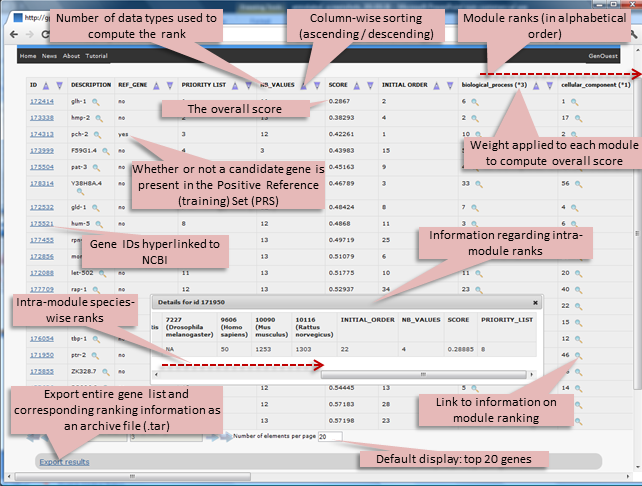
The weight used in each module to compute the overall score is indicated in brackets. Any unrecognised Entrez Gene IDs are indicated by “NOT FOUND!” and no ranks are returned. Redundant genes in the candidate list are reported only once in the results page. The output list is ordered (prioritized) according to the overall score; it can be reordered based on the ranks of individual modules. Information regarding the intra-module ranks (initial order, NB_values, score, priority list and the species-wise ranking) is accessible through the magnifying glass icon. The table in the html output displays the top 1000 genes; the entire gene list and corresponding ranking information can be exported as an archive file (.tar) via the “Export results” link at the bottom of the page.
The Results archive file (.tar) consists of a main ‘results.out’ file with the prioritized gene list and module-wise ranking similar to the html output. Extended ranking information is available through the output files for each module. In addition to global ranking and module scores, species-wise ranks are listed (NCBI Taxonomy IDs (e.g. Human = 9606) indicate the species that contributed to the ranking). The welcome page includes a link to a brief tutorial for GPSy.
Contacts
If you need help, please contact Contact GenOuest. For questions concerning the design of GPSy, please contact Frederic Chalmel.
